algorithmic modeling for Rhino
Protein Database Downloader
The PDB plug-in was rather inconvenient in requiring your to manually download Protein Database Files and it lacked an atom radius output, so I discovered Python has a built-in http download library, and I added atomic radii from Wikipedia.
The new Cocoon marching cubes plug-in along with Kangaroo MeshMachine to refine the mesh better than the native tweaky Cocoon refine component are included.
The smaller example takes about 15 seconds. The huge one a minute or two.
Just enter the PDB symbol, and you can browse for those here:
http://pdb101.rcsb.org/motm/motm-by-title
The small one above is a DNA repair enzyme wrapped around DNA:
http://www.rcsb.org/pdb/explore.do?structureId=3UGM
Some are just too big to test and will likely blow up Cocoon or at least MeshMachine.
I thought these structures might be useful to 3D printer enthusiasts looking for fun blobby shapes to build with.
Views: 6663
Replies to This Discussion
-
I would like to think changing the DNA of our thinking brains might be the key to our Survival.
I was interested to learn 99 Million years ago 2 ants Supposedly were locked in mortal combat which was found in Burmese Amber.
http://news.discovery.com/animals/insects/ancient-ants-in-amber-wer...
Fast forward 99 Million years, and we are still kill each other, with our very own thinking brains! Nothing seems to have changed in 99 million years. Yet we have the audacity to think we now going to solve Global Warming? conveniently renamed Climate Change!
are you kidding me! -
-
Just literally google "is world getting better" and check up some statistical data. Unless you are a conspiracy theory enthusiast XD, you will see that world is getting better, slowly but steadily.
Anyway, hope you find this info reassuring and believable (some people just enjoy boiling in the negativity) and have a good and productive day.
-
-
Hey, where can I download marching cubes components?
-
-
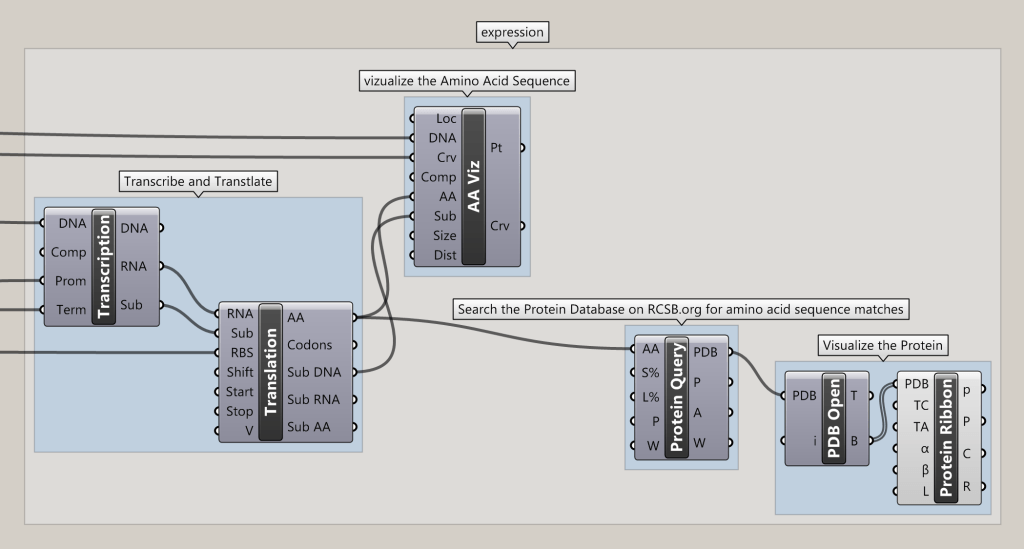
This looks great, Nick. Thanks for sharing. You may also be interested in the suite of Grasshopper tools called "Alba" that I've just release for synthetic biology. It i
ncludes a component that allows you to search an amino acid sequence on RCSB, download the best match, and visualize the structure of the protein as a "ribbon" showing beta sheets and alpha helices. You can find more info and download it here.
Cheers,
Ryan
-
-
Ribbons! Fun candy.
I'd also like to surface electron microscope surface files, not just solved molecule ones:
-
-
...also, PDB models often lack the full often radially symmetrical assembly, only giving one unit, and I don't know how to recreate the full protein machine like the PDB page often indeed shows fully assembled.
-
-
Today's highlight was a heat shock protein cluster, for instance, but loading it into iMolview only gives one unit:
-
-
Ryan, your download link is failing for both your plug-ins:
"Error: INSERT INTO users (first, last, email, field, comment, date, timestamp) VALUES ('Nik', 'Willmore', 'nikwillmore@gmail.com', '', '','2016-04-29 20:39:48 Friday','1461976788' )"
-
-
Thanks for letting me know, Nick. It is fixed now.
-
-
New problem: the Protein tab is missing that I see here in your screenshot, but the rest is intact.
http://www.ryanhoover.org/images/Alba_Demo_GFP_DNA_TxTl_ProteinQuer...
Tried with both latest Rhino/Grasshopper and Rhino 6 WIP. No files were blocked in Windows. Using 64 bit Windows 7. Tried placing folder in User Objects as well as individual folder contents. There are 43 files, which is indeed 4 files short of how many components you show.
I noticed one sample file also required gHowl and also installed that.
-
- ‹ Previous
- 1
- 2
- 3
- Next ›
© 2024 Created by Scott Davidson.
Powered by
![]()